Notre laboratoire a été reconnu comme unité de recherche commune de l'INSERM et de l'université d'Aix-Marseille en 2008, sous la direction de Catherine Nguyen. Il est situé sur le campus de Luminy et n'a cessé de croître, passant de 30 personnes en 2008 à 68 aujourd'hui. À l'aide de la génétique, de la génomique et de la bio-informatique, nous cherchons à déchiffrer les mécanismes biologiques complexes impliqués dans le fonctionnement physiologique des organismes, ainsi que ceux conduisant à des pathologies chez l'homme telles que les hémopathies malignes, la septicémie, le paludisme ou les maladies cardiaques. Pour développer un tel projet, notre laboratoire rassemble des chercheurs et des ingénieurs possédant un large éventail de compétences (biologie cellulaire et moléculaire, génétique, génomique, bio-informatique), convaincus que la combinaison d'approches expérimentales et bio-informatiques permettra l'émergence de nouvelles connaissances et de nouveaux concepts. L'activité de recherche bénéficie de nos installations de génomique (TGML) et CRISPR.
Notre projet de recherche s'articule autour de deux axes :
| génétique et génomique des maladies multifactorielles |
Le premier axe est consacré au déchiffrage de l'organisation fonctionnelle du génome, à la caractérisation du rôle fonctionnel des ARN non codants, à l'analyse des réseaux d'interactions moléculaires par la génomique et la bio-informatique, et au développement de nouvelles stratégies pour intégrer des informations biologiques hétérogènes dans des modèles théoriques prédictifs. Le deuxième axe vise à caractériser les profils génétiques, transcriptomiques, épigénomiques ou protéomiques associés aux hémopathies malignes, à la septicémie, au paludisme ou aux cardiomyopathies, afin d'identifier les éléments et réseaux régulateurs perturbés par ces pathologies, ainsi que les biomarqueurs et cibles thérapeutiques pour ces pathologies.
Notre laboratoire est volontairement tourné vers l'interdisciplinarité et la formation. Nous menons non seulement des projets de recherche fondamentale, mais aussi des projets collaboratifs avec des hôpitaux et des entreprises de biotechnologie. Cette politique s'inscrit pleinement dans le cadre du plan stratégique de l'INSERM. De plus, nous sommes fortement impliqués dans la formation universitaire, du niveau licence au doctorat.
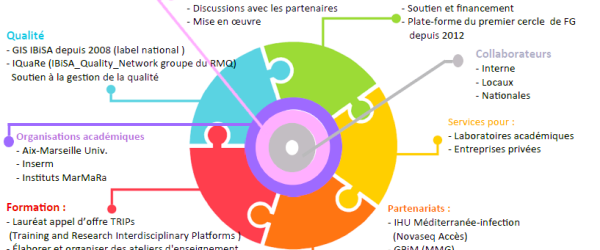
Notre laboratoire développe de nombreuses interactions avec les laboratoires de l'Université d'Aix-Marseille. Nous participons à des recherches dans les disciplines considérées comme prioritaires par le comité scientifique de l'université (génétique, immunologie, cancer) et celles menées par les services hospitaliers universitaires d'immunologie, de reproduction et de neurosciences. De plus, notre laboratoire est membre des instituts interdisciplinaires Marmara (génétique), ICI (onco-immunologie) et CenTuri (http://centuri-livingsystems.org/), qui ont pour objectif de déchiffrer la complexité de la vie à l'aide d'approches interdisciplinaires.
| Recherche au TAGC |